Breadcrumb
- Home
- Research
Research
Research Interests
The accumulation of somatic mutations in an estimated four to seven key genes is necessary for the development of most human cancers. These mutations lead to aberrant cell proliferation by activating proto-oncogenes and inactivating tumor suppressor genes. Consequently, factors increasing the cellular mutation rate accelerate the onset of cancers; those decreasing the mutation rate delay it. Thus, understanding how mutations arise is important to understanding carcinogenesis and to developing novel approaches to prevent it.
Our laboratory studies the mechanisms by which mutations occur in eukaryotic cells. It turns out that most mutations result from the replication of damaged DNA. DNA damage is problematic because it blocks replication by classical DNA polymerases – those involved in normal DNA replication and DNA repair. Without a means of overcoming these replication blocks, cells cannot divide. For this reason, cells possess several non-classical polymerases capable of replicating through DNA damage. These non-classical polymerases are highly error-prone and are responsible for them is incorporation events that ultimately give rise to the mutations.
Our long-term goal is to understand the mechanisms of non-classical DNA polymerases and various replication accessory factors involved in mutagenesis. We use a variety of approaches including biochemical studies (ligand binding studies, enzyme activity assays), structural biology (X-ray crystallography, X-ray scattering), and cell-based techniques (yeast genetics, human tissue culture). We hope that this work will contribute to our understanding of the origins of mutations and cancers and perhaps gain new insights into their prevention.
Projects
Investigating the mechanism and composition of the TLS multi-protein complex

Translesion Synthesis (TLS) is a DNA damage bypass pathway which requires numerous proteins to come together in a dynamic, mulit-protein complex. We seek to understand the TLS complex through studies utilizing single molecule TIRF assays. Previously we have determined that PCNA can form both PCNA "tool-belts" and "bridges" in the ternary complex containing PCNA and two TLS polymerases, Rev1 and Polymerase eta. Our goal is to use these same techniques to discover more about the mechanism and architecture of the TLS multi-protein complex. https://www.ncbi.nlm.nih.gov/pubmed/27325737
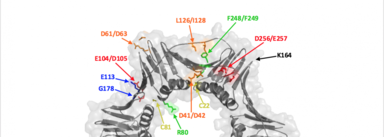
PCNA separation of function mutations
PCNA serves as a central component of many DNA repair processes, including mismatch repair, break-induced replication, error-free bypass, and TLS. Our lab has solved the structures of numerous separation of function mutations relating to each of these processes in order to determine how these mutations may affect the function of PCNA. https://www.ncbi.nlm.nih.gov/pubmed/?term=Todd+Washington+PCNA+mutations+structure http://www.rcsb.org/pdb/results/results.do?tabtoshow=Current&qrid=B9F4F0C9
The role of PCNAmonoubiquitylation and SUMOylation in TLS
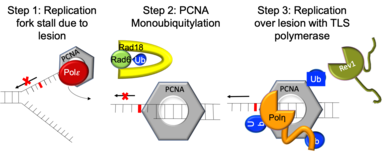
A key step in the TLS pathway is the monoubiquitination of proliferating cellular nuclear antigen (PCNA). Ubiquitination is accomplished by a ubiquitin ligase complex Rad6-Rad18 (Rad6-18), and is stimulated by one of the specialized TLS polymerases, polymerase eta (Pol eta). However, despite the importance of this essential step, it is unclear whether PCNA monoubiquitination by Rad6-18 results in recruitment of TLS polymerases or stimulation of TLS polymerase enzymatic function. Likewise, PCNA SUMOylation in yeast appears to have a role in TLS as well by recruiting Rad6-18. Our goal is to study the role that each of these PCNA modifications plays in TLS.